News
December 17, 2015
By Nuritas
Tracking the growth of the gut microbiota and what this means for your health

December 17, 2015 – Nuritas
Contributing Authors: Dr. Dunja Skoko and Meaghan Lee-Erlandsen
The precise measurement of microbial populations in the gastrointestinal tract could be key in identifying novel therapies for Irritable Bowel Syndrome (IBS) and other conditions in the gut.
More and more scientific research is confirming that the overall health of our bodies largely depends on the health of our gut. A healthy gut is hard to achieve however without a perfectly balanced intestinal flora. Flora consists of beneficial bacteria that help us digest our food and fight outside offenders and it is important for our bodies to keep this intact. Indeed, any disturbance of the beneficial mutual relationship between intestinal microbes and the body is suggested to play an important role in numerous intestinal conditions and diseases. For example, antibiotic use, known to disturb the flora, may cause stomach discomfort and may predispose individuals to Irritable Bowel Syndrome (IBS). The opposite is also true in that the alteration of the bacteria in the gut with the use of probiotics may improve the gut flora, helping to decrease the symptoms associated with IBS. Unfortunately, it was not possible to directly observe growth rates of the intestinal inhabitants and evaluate and track their contribution to the onset or alleviation of intestinal disorders until now.
A disruptive and promising method in understanding and managing gastrointestinal health has emerged from the Wyss Institute for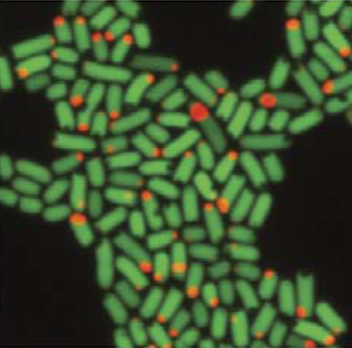 Biologically Inspired Engineering at Harvard University and Harvard Medical School.
Biologically Inspired Engineering at Harvard University and Harvard Medical School.
According to the new report published in Nature Communications, scientists can now, for the very first time, accurately measure population growth rates of the microbes that live inside mammalian gastrointestinal tracts. The new method has been named “distributed cell division counting,” or DCDC.
The team used fluorescently tagged E. coli cells to measure microbial growth rates of 10 consecutive generations on their journey through the digestive tract. Interestingly, DCDC shares conceptual similarities with mark and recapture methods employed by ecologists to measure the growth rates of animal populations in the wild. Namely, individual animals are marked, released to the wild and later their offspring is resampled for the mark which allows estimation of the whole population growth dynamic. Similarly, DCDC uses a microbial scale mark and recapture strategy: naturally occurring E. coli cells isolated from the gut are modified to express fluorescently labelled proteins that self-assemble to form a stable bright particle in each bacterial cell. The bright particle is observable by fluorescent techniques such as microscopy. The fluorescent bacteria is fed to mice and the presence of tagged microbes is then evaluated within the faeces in regular time intervals. Counting the number of exiting bacteria allows the researchers to estimate the complex population dynamics in the gut.
According to Cameron Myhrvold, the lead author of the published study, “Many different approaches to managing gastrointestinal health — such as the use of antibiotics, probiotics, genetically engineered bacteria, and therapeutics — rely on us being able to tell how these treatments affect the growth, division and death of certain microbial populations in our guts,”.
This new method is a promising scientific and diagnostic tool that has a very high potential to reveal both the secret life of our intestinal inhabitants and to give us a better insight into how these invisible inhabitants contribute to our health and possibly many of our diseases.
We at Nuritas™ look forward to contributing to this important area in the future and providing easy and cost-effective access to natural ingredients that will help people manage the healthy balance of their gut and overall health.
Reference:
Myhrvold, C. et al. A distributed cell division counter reveals growth dynamics in the gut microbiota. Nat. Commun. 6:10039 doi: 10.1038/ ncomms10039 (2015)





 Previous
Previous
